Abstract
Recombinant Saccharomyces cerevisiae (yeast) represents a unique and attractive vehicle for delivering antigens in cancer or infectious disease vaccine immunotherapy protocols, as it has been shown to be extremely safe and can be administered multiple times to hosts. In the studies reported here, we describe the effects of recombinant yeast treatment on immature murine dendritic cells (DCs). Yeast expressing human carcinoembryonic antigen (CEA) was studied as a model antigen.
Injection of mice subcutaneously with yeast-CEA resulted in a rapid increase in MHC class II (+) cells and total antigen-presenting cells in the draining lymph nodes. Following yeast-CEA treatment, DC rapidly elevated both MHC class I and class II, numerous costimulatory molecules, and other markers of DC maturation, and secreted a number of types I inflammatory cytokines.
Expression Arrays Gene synthesis also revealed the rapid upregulation of numerous cytokine and chemokine mRNAs, as well as genes involved in signal transduction and antigen uptake. Functional studies demonstrated increased allospecific reactivity of DCs after treatment with yeast-CEA or control yeast.
Furthermore, treatment of DC with yeast-CEA resulted in specific activation of CEA-specific CD8(+) T cells in an MHC-restricted manner in vitro. Lastly, vaccination of CEA transgenic mice with yeast CEA elicited antigen-specific CD4 (+) and CD8 (+) immune responses in vivo. Therefore, these studies taken together form a scientific justification for the use of recombinant yeast in vaccination protocols against cancer or infectious diseases.
Results
Whole Genome Sequence Analysis
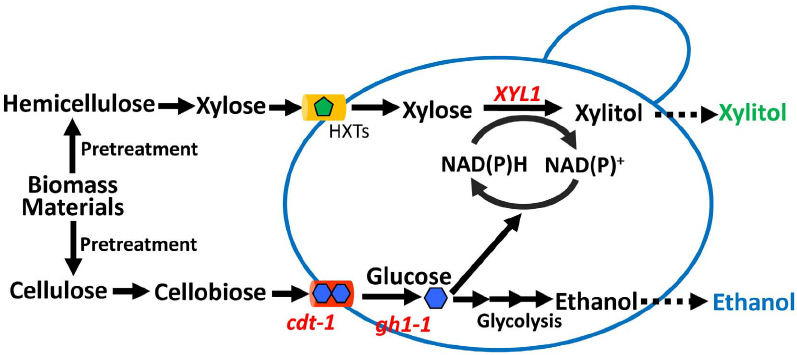
Recently, we reported the development of an industrial D-xylose using a strain of S. cerevisiae, GS1.11–26, using a combination of metabolic engineering, genome shuffling, and evolutionary adaptation. Briefly, we integrated the XylA gene encoding xylose isomerase from the bacterium Clostridium phytofermentans along with all known genes important for D-xylose and L-arabinose metabolism into an industrial bioethanol production strain, ER, (Ethanol Red).
However, the recombinant strain designated HDY.GUF5 was unable to utilize D-xylose. We then mutagenized this strain by EMS and selected a mutant isolate M315 that could grow slowly on D-xylose but had poor D-xylose fermentation ability. Genome shuffling of this mutant with its parent HDY.GUF5, followed by selection for faster D-xylose growth, resulted in little improvement. Subsequently, we performed the evolutionary adaptation in serial batch cultures containing D-xylose as the main carbon source.
A surprising observation during this adaptation process was a dramatic increase in the fermentability of D-xylose in the second serial batch culture. We proposed that a crucial genetic change had occurred at that step, resulting in a rapid gain in performance. To elucidate this change, we sequenced the genome of the original parental strain HDY.GUF5 and the final evolved strain GS1.11–26 and performed a global genome sequence comparison.